27 February 2024
I am writing to you regarding your recent call for feedback on the manuscript titled “Aluminum and Amyloid-in Familial Alzheimer’s Disease” by Mold and colleagues [1] that you have submitted on PubPeer (https://pubpeer.com/publications/1F23A5E2AE061A909F4D46F4919B50).
Although this manuscript nicely follows up on the previous publications by the research group that investigated previously on the quantification of aluminum in the brain of healthy patients and patients suffering from Alzheimer’s disease and related dementia (ADRD) [2-4], it also shows signs of the same methodological flaws and limitations that could be observed in their previous publications.
I would therefore provide my concerns on the methodology, results, and subsequent conclusions made by the authors.
1. Sampling type: In this article, the authors mentioned the following sources: “Brain tissues, frozen and fixed, from donors who carried the PS1-E280A mutation were obtained from the brain bank of the Universidad de Antioquia, Medellin, Colombia and research was carried out following ethical approval by Keele University (ERP2391). The consultant neuropathologist at the London Neurodegenerative Diseases Brain Bank chose the control brain tissues for us and their detailed data are presented elsewhere (Exley and Clarkson, unpublished)”. These samples must originate from frozen tissue samples in place of formalin-fixed samples, which provides an improvement from formalin-fixed brain samples from their previous publications. However, the omission of providing data from control samples performed in parallel with a familial form of Alzheimer’s disease (FAD) samples, while omitting direct access to such data (quote “their detailed data are presented elsewhere (Exley and Clarkson, unpublished)” deviate from common practice in scientific rigor. The authors should have included this information, including a primary source to access such information, to allow the readers to appreciate the values reported in this manuscript.
Based on the information provided in the Supplementary Figure 2 of this article, I suspect that the source of such controls, based on the number of biological samples (n=20) must be their study published in Scientific Reports in 2020 [3].
2. Analytical method: The authors have utilized the transversally heated graphite furnace atomic absorption spectrometry (TH GFAAS). This method has been intensively used by the authors in several of their publications [2, 4-9], and would therefore consider this technique as a reliable technique for the quantification of aluminum. However, their previous study has highlighted several issues in terms of validation, as precedent samples have highlighted significant intra-sample variability within technical replicates, as illustrated with the data presented in their Metallomics study (Fig. 1) [9]. In this study, they measured aluminum values (as microg/g dry tissues) in 60 brain donors in different brain regions (frontal, occipital, parietal, and temporal). In several instances, they have reported negative values, which would indicate values below background noise, which raises an issue in terms of accuracy and validation of the method. We cannot exclude that we may have some heterogeneity in the deposition of aluminum within sampling sites of the same sample tissue. However, the minute amount of tissue (0.6-2 g, according to the authors) sampled makes this assertion unlikely and may suggest serious limitations in the analytical method employed.

Figure 1. Representative distribution of technical replicates (1-3 replicates/biological samples) amongst 60 donors in four different brain regions (frontal, occipital, parietal, and temporal lobes). The mean and standard deviation are depicted in red, donor sources are listed as numbers.
Reproducibility is a key element of scientific rigor and the intrinsic lack of validation procedure of the method as commonly performed with other analytical instruments (e.g., high-performance liquid chromatography, liquid chromatography with mass-spectrometry) in terms of linearity, accuracy, precision, range, detection, and quantification limits is paramount to ensure this analytical method as accurate. One key issue we have observed is that the authors appear not capable of reproducing their findings between samples of the same category in their studies [9, 10] (Fig. 2).
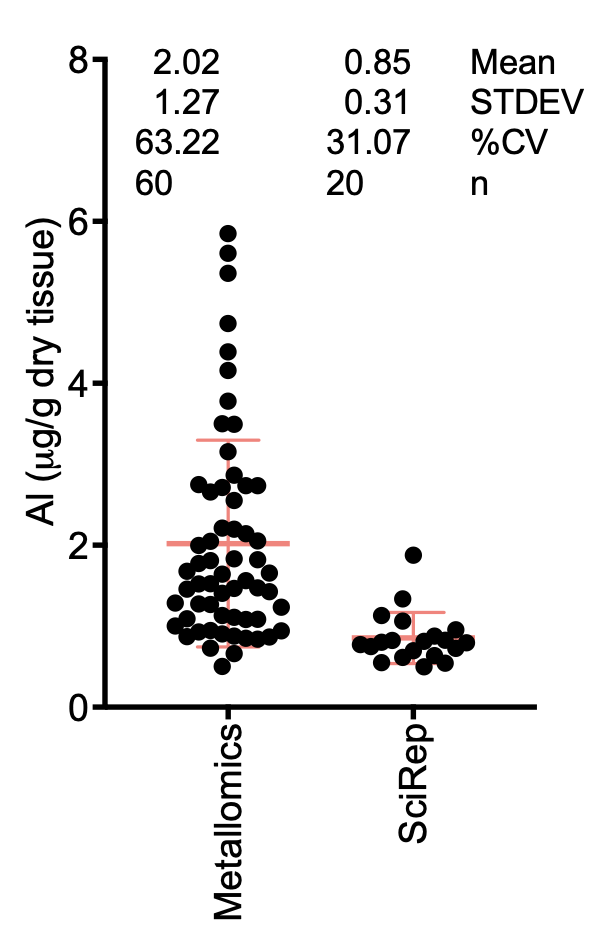
Figure 2. Raw measurement of average brain concentrations between two control batches from two independent studies, using the same analytical method and same research group. For each biological sample, the biological average was obtained from technical replicates (excluding negative and missing values) for each brain region, while the brain average was obtained using the biological average of each region (frontal, occipital, parietal, and temporal). Note the huge difference (p<0.0001, two-tailed, Mann-Whitney test) between the two cohorts of controls and the important coefficient of variation observed within each sample.
This is problematic, as we expect that the utilization of the same instrument, by the same laboratory would alleviate variability in such samples.
In this study [1], out of 12 patients with AD, the authors could only achieve three technical replicates in less than 50% of the analysis, with an average coefficient of variation between 41% and 57%, which is a magnitude much higher than the average 5% error tolerance we would expect for a precise measurement in analytical method. I consider this important margin error and the incapacity to consistently obtain 3 technical replicates as serious limitations and caveats for a technique considered as precise and reproducible.
3. The statistical inaccuracy of considering technical replicates as biological replicates. Technical replicates are important replicates needed to increase the accuracy of a measurement. However, technical replicates cannot be considered as independent populations, a key condition in statistical analysis involving comparisons of variances (e.g., t-tests, one-way analysis of variance), in which the independence of the data (in this case individuals) is needed. The exact and accurate numbers of technical replicates remained vague. In the Table 1 legend, the authors mentioned 1-4 replicates, yet the technical replicates are highly variable between brain regions of the same patient (e.g., #case 221, number of technical replicates (n) per tissue: occipital n=1; temporal n=2; frontal n=2; parietal n=3). It would have been more appropriate for the authors to report this measurement as average for each tissue sample.
4. The importance of displaying raw data: In this study, the authors significantly deviated from their previous results representations. I consider that representing the raw data (after noise correction or normalization against total protein load) with the least amount of manipulation is the most accurate representation of data (Fig. 3).
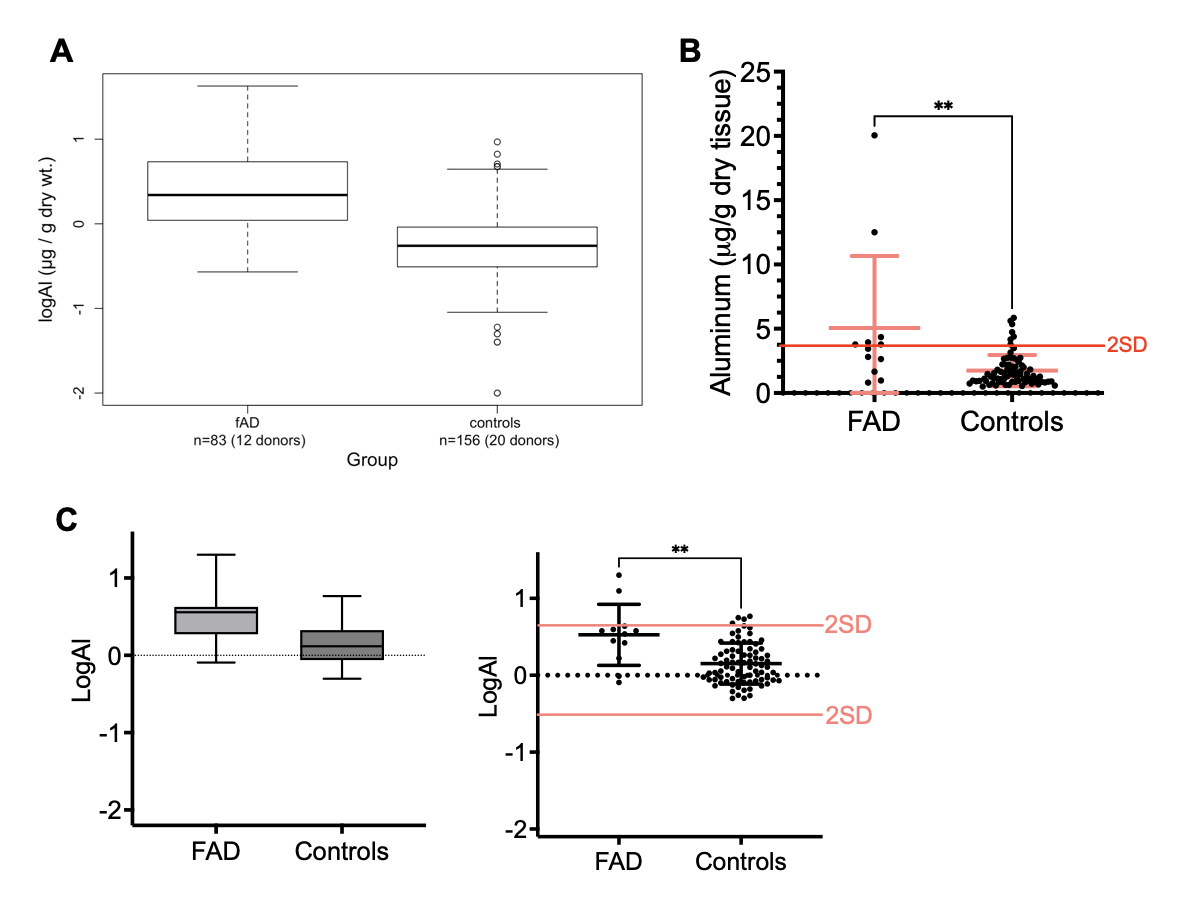
Fig. 3. Comparison between FAD and controls groups. A) Original data representation as presented in the study. B) Raw brain average values between FAD patients and controls (n=60+20). Mean and standard deviation bars are highlighted in red. Note the statistical significance obtained (p<0.01, two-tailed t-test, Mann-Whitney correction), which also align with the 2SD variation. C) Data representation in log values in both box-plots and dot-plot representations.
This is problematic, as we expect that the utilization of the same instrument, by the same laboratory would alleviate variability in such samples.
In this case, I consider representing the amount of aluminum as µg/g dry tissue the most accurate and direct representation of the data. In this study, the authors decided to represent their data distribution compared to controls as log Al (log microg/g). The representation of such values as log10 values is in my opinion unsubstantiated, and I am concerned that the adoption of such representation was chosen to camouflage the significant variability in the data. Homoscedasticity is a key element in statistical analysis between groups and the important variability from both the technique (see %CV) and between patient samples raises concern about the statistical relevance regarding elevated aluminum in fAD patients compared to control groups.
Nevertheless, we cannot ignore the statistical difference in brain aluminum content observed in FAD patients compared to control patients, however, such samples must be age-matched and certainly will benefit additional studies to improve the statistical power of analysis to confirm the elevation of brain aluminum in FAD patients.
5. Microscopy methods: This remains a methodological flaw that has been carried over by the group over the years and remains problematic. Firstly, the use of brain tissue, especially aged brain tissue carries the risk of autofluorescence inherent to the accumulation of lipofuscin [11]. Lipofuscins are pigmented granules, that can be present in lysosomes that are known to display autofluorescence with emission wavelength ranging from 420 to 700 nm, with peak emission occurring in the 570-600 nm range. It is therefore important that such autofluorescence is accounted for by the subtraction of non-specific fluorescence (e.g., setting the exposure on unstained tissues), or the use of narrow optical filters to avoid the detection of unwanted parasitic fluorescence signals. This is unfortunately not accounted for as the authors have used two cubes (U-MNIB3 and U-MWBV2) that do not provide discrimination in the emission wavelength, providing this green-yellow tint observed on the micrograph pictures (Fig. 4).
This issue is also becoming highly problematic, as the use of Lumogallion as a fluorescent probe for aluminum, displays an emission spectrum overlapping with lipofuscin (with peak emission around 580 nm as well). Such overlap is highly problematic as it does not allow the experimenter to distinguish autofluorescence from the tissue to the fluorescence specific to the Lumogallion-aluminum complex. The absence of controls to remove such background noise is highly problematic and limits the interpretation of the results displayed in this study. In addition, the lack of inclusion of controls (not fAD donors) is again a serious concern when it comes to scientific rigor, as these negative controls are necessary to validate the presence of an increased fluorescence signal inherent to the presence of aluminum. In addition to the fluorescence techniques and instrument employed, I consider the claim of being able to identify cell types (e.g., glial cells, as the authors claim in Fig. 5) without having the appropriate fluorescent counterstaining and use of cell markers (e.g., S100β or glial fibrillary acidic protein) to be bold and unsubstantiated. Even trained eyes of neuropathologists would require the use of particular histological stains to speculate on cell types in the brain, even less by looking at an unstained brain sample microsection.
Finally, I consider the use of thioflavin-S as staining for amyloid plaques would have greatly benefited from a combination of an antibody-based staining (using antibodies raised against human Aβ1-42) to demonstrate the validity of the staining protocol, but also to complement and demonstrate that the immunofluorescence of Aβ was remarkable with the one from the Lumogallion staining.
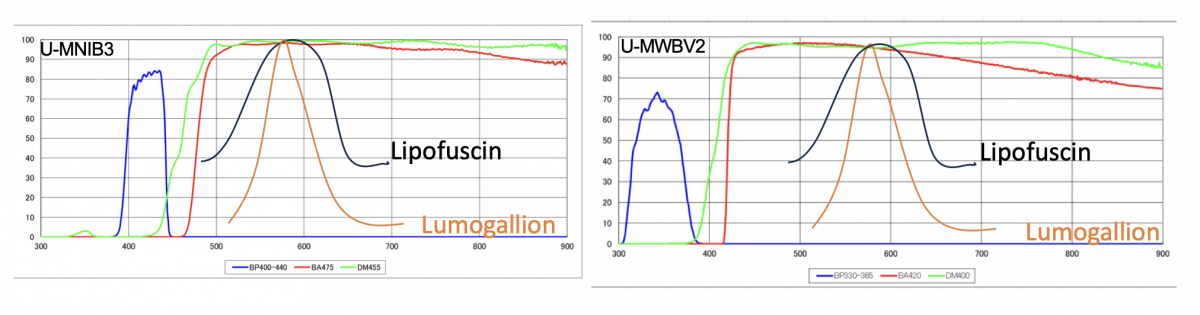
Fig. 4. Schematic representation of the excitation and emission transmission range of the microscope cube filters. Approximate emission spectra for lipofuscin (black) and Lumogallion (orange) were superposed. Note the spectral overlap of both dyes, which is hampered by the discrimination in emission range intervals towards a specific wavelength above 480 and 420 nm respectively.
In conclusion, this manuscript, as interesting as it is and revivifies the aluminum hypothesis in the pathophysiology of Alzheimer’s disease, remains deeply flawed with the same limitations and methodological flaws that were highlighted in a previous commentary (https://healthfeedback.org/claimreview/vaccines-do-not-cause-neurologica...).
The serious limitations and inconsistencies of this study with the previous publications from the same group in regards of aluminum depositions in human brains by both quantitative and qualitative approaches remain questionable, and I do not share the bold and confident interpretations and conclusions stated by the authors in the discussion.
However, we cannot dismiss the observations done (and that appear consistent between the studies) about an increased level of brain aluminum in patients suffering from ADRD, regardless of its origin (sporadic or familial forms). Several hypotheses could explain these observations and we can speculate that impaired aluminum transport across the blood-brain barrier (BBB) may occur. Two possible entrance paths at the BBB for aluminum occur: either a passive paracellular diffusion inherent to a weakening of the barrier by chronic exposure to Aβ peptides or by mutated PSEN1 [12, 13], or an indirect consequence of impaired iron metabolism (abnormal brain transferrin uptake at the BBB) resulting in ferroptosis [14].
Until more rigorous and different approaches are brought forward to validate these findings, further studies are needed to support the aluminum hypothesis of aluminum in ADRD. However, the statement of aluminum being a causing agent of ADRD as claimed by the corresponding author over the years remains to be supported by robust and compelling evidence, to firmly establish a strong causation as of a weak correlation.
Abraham Al-Ahmad
Ph.D., Associate Professor
Graduate Program Director
Conflict of interest
The author has no conflict of interest to report.
References
[1] Mold M, Linhart C, Gomez-Ramirez J, Villegas-Lanau A, Exley C (2020) Aluminum and amyloid-beta in familial Alzheimer's disease. J Alzheimers Dis 73, 1627-1635.
[2] Mirza A, King A, Troakes C, Exley C (2017) Aluminium in brain tissue in familial Alzheimer's disease. J Trace Elem Med Biol 40, 30-36.
[3] Exley C, Clarkson E (2020) Aluminium in human brain tissue from donors without neurodegenerative disease: A comparison with Alzheimer's disease, multiple sclerosis and autism. Sci Rep 10, 7770.
[4] Exley C, House E, Polwart A, Esiri MM (2012) Brain burdens of aluminum, iron, and copper and their relationships with amyloid-beta pathology in 60 human brains. J Alzheimers Dis 31, 725-730.
[5] Shardlow E, Linhart C, Connor S, Softely E, Exley C (2021) The measurement and full statistical analysis including Bayesian methods of the aluminium content of infant vaccines. J Trace Elem Med Biol 66, 126762.
[6] Mold M, Umar D, King A, Exley C (2018) Aluminium in brain tissue in autism. J Trace Elem Med Biol 46, 76-82.
[7] Mold M, Chmielecka A, Rodriguez MRR, Thom F, Linhart C, King A, Exley C (2018) Aluminium in brain tissue in multiple sclerosis. Int J Environ Res Public Health 15, 1777.
[8] Exley C, Vickers T (2014) Elevated brain aluminium and early onset Alzheimer's disease in an individual occupationally exposed to aluminium: a case report. J Med Case Rep 8, 41.
[9] House E, Esiri M, Forster G, Ince PG, Exley C (2012) Aluminium, iron and copper in human brain tissues donated to the Medical Research Council's Cognitive Function and Ageing Study. Metallomics 4, 56-65.
[10] Martinez CS, Uranga-Ocio JA, Pecanha FM, Vassallo DV, Exley C, Miguel-Castro M, Wiggers GA (2022) Dietary egg white hydrolysate prevents male reproductive dysfunction after long-term exposure to aluminum in rats. Metabolites 12, 1188.
[11] Yu N, Su S, Cui H, Fang Y, Sun J, Cao Y, Ma C (2021) Autofluorescence in human tissue really matters. Pain 162, 2780.
[12] Raut S, Patel R, Pervaiz I, Al-Ahmad AJ (2022) Abeta peptides disrupt the barrier integrity and glucose metabolism of human induced pluripotent stem cell-derived brain microvascular endothelial cells. Neurotoxicology 89, 110-120.
[13] Raut S, Patel R, Al-Ahmad AJ (2021) Presence of a mutation in PSEN1 or PSEN2 gene is associated with an impaired brain endothelial cell phenotype in vitro. Fluids Barriers CNS 18, 3.
[14] Campos-Escamilla C (2021) The role of transferrins and iron-related proteins in brain iron transport: applications to neurological diseases. Adv Protein Chem Struct Biol 123, 133-162.
Comments
Comment
Much of the concern expressed by Al-Ahman revolves around the use of tissue specimens for assessing aluminum accumulation and the imprecision of the analytical measurements. Tissue has always been recognized as an appropriate specimen for assessing body burden of metals but has the disadvantage of being heterogeneous and, hence, two samples located close to each other often give widely divergent results. Due to this reason, biological fluids, rather than tissue, are most commonly used to assess body burden of metals but unfortunately this often does not reflect body burden. A good example is zinc where plasma or urine analysis can provide accurate and precise measurements that have little bearing on assessing body burden. Tissue analysis gives much better results but side by side samples often to not agree. Also of course obtaining a tissue specimen is unpleasant for the patient and phlebotomy or urine collection is much simpler. Lymphocyte analysis has been proposed as a compromise and has shown much promise, but I do not think it is widely used at the present time especially since zinc deficiency is not a major health issue. When aluminum was first shown to be a major health issue in hemodialysis patients it was important to standardize the analytical assessment of body burden of aluminum. Serum measurements had their limitations, and tissue and bone analysis was proposed and used in a few dialysis centers. Fortunately, serum or plasma did provide a reasonable means of assessing body burden and are now the specimen of choice. Lymphocytes were also considered but did help much and I do not think offered any advantages over serum or plasma measurements. Obviously, the only specimen which can be used to assess aluminum accumulation in the central nervous system is brain tissue, which requires extensive processing prior to the final measurement. It appears that Exley recognized this, and he and his coworkers did the best they could at trying to show that aluminum accumulation does occur in Alzheimer's disease affected patients. The more brain tissue that is available then the precision of the measurement would improve, but it appears that the amount of brain tissue available to the Exley group was limited. Despite the limitations of the use of brain tissue and its heterogeneity, it appears to me that valuable information was forthcoming from the study and deserved to be published. Perhaps, however, Dr. Exley could have been less dogmatic in expressing his conclusions that aluminum does cause Alzheimer's disease. Al-Ahman expresses some concern about the precision of the electrothermal atomic absorption technique. This is a tried-and-true technology that was developed in the 1960s and first applied to the analysis of biological specimens in 1973. It is both an accurate and precise technique and Exley and his group have had extensive experience with the analytical system. There are newer techniques but are not without their drawbacks. The use of mass spectrometry for the final measurement also has problems with polyatomic interferences which require high resolution mass spectrometry to obtain the best data. Yes perhaps Dr. Exley was somewhat dogmatic about stating definitively that aluminum and Alzheimer’s disease are intertwined, but it could be pointed out that those neuroscientists who support the Aβ hypothesis are equally dogmatic about their confidence that Aβ accumulation is central to the development of Alzheimer's disease. How many thousands of times has this been stated over the past 40 plus years. Countless millions of dollars have been expended on drug development and treatments that still are of little value. Since aluminum is so highly abundant on Planet Earth, we will probably never know if this element is the primary cause of the disease or perhaps if the biochemical and neuropathological abnormalities observed in the disease just happen to possess affinity for aluminum. This would not be surprising since the toxic aluminum species Al3+ is highly reactive and has a high affinity for phosphorylated proteins. One feature of the aluminum hypothesis that provides support for aluminum playing an important role in the development of Alzheimer’s and related disorder is that most of the biochemical and neuropathological changes seen in Alzheimer’s disease can be reproduced in experimental animals. Very few neuroscientists understand the complex chemistry of aluminum, yet this is central to designing meaningful experiments. Additionally, the selection of appropriate animal model systems is extremely important, and the use of rabbits is an obvious choice since they more closely resemble primates than rodents [1]. With this animal model system and using aluminum maltolate as the preferred aluminum compound remarkable similarities between Alzheimer's disease patients and the changes seen in rabbits, are observed. This is an acute exposure to the animals and requires the intracerebral administration of aluminum maltolate. Comparison of the changes in Alzheimer’s disease and the acute exposure in rabbits is summarized in the following Table. There is a remarkable similarity with the exception that paired helical filaments in the animal model are not observed, although there are intraneuronal neurofilamentous aggregates that are positive for hyperphosphorylated tau. It could be argued that once these intraneuronal neurofilamentous aggregates are in place then a structural rearrangement could occur with time—perhaps months or years. 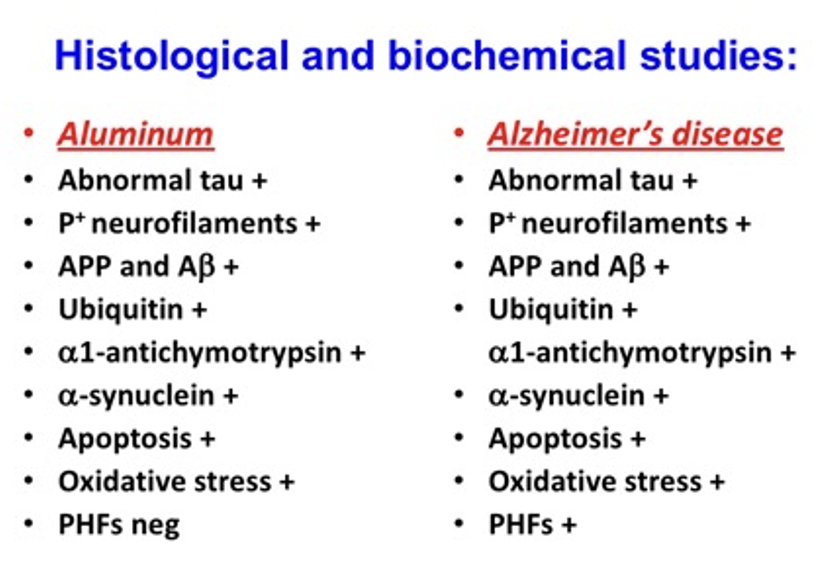
Imagine what interesting studies could have been forthcoming if a fraction of the funding for Aβ research could have been redirected. Yet the only argument against the aluminum hypothesis is that “Everyone knows that aluminum does not cause Alzheimer’s disease”. This is widely stated by the vast majority of physicians/neuroscientists and, of course those working for the best interests of the aluminum industry. This is despite the data shown above and the fact that aluminum is the most abundant metal in the earth’s crust and the third most abundant element. Fortunately, aluminum is locked in complex inorganic and organic species making it relatively innocuous except perhaps in certain individuals and also as a result of the aging process. Good science would undoubtedly have contributed a great deal to such a field of research.
On a personal note, in my old age I see memory loss in members of my own family and I greatly resent the anti-aluminum faction of our society led by the aluminum industry who have spearheaded the charge to stifle any research that might affect the image of aluminum. Dr. Exley has made many excellent contributions to carry the banner of the aluminum hypotheses, not least of which are the international conferences that he has organized in memory of his mentor, Dr. Derek Birchall.
I hope what I have written is helpful to you in assessing what to do about the PubPeer submission. I had not heard of PubPeer until I received your email, but it sounds like an interesting approach to stimulating discussion especially concerning controversial topic. Some of my comments are just a means of letting off steam but so much could have been accomplished to understand what could be an important line of research to help those with this dreadful neurodegenerative disease. I have written most of the above from memory. Perhaps in my old age I have been spared the consequences of brain aluminum accumulation—I am 88 years old which puts me in a vulnerable age group for Alzheimer's disease.
John Savory
Reference
[1] Graur D, Duret L, Gouy M (1996) Phylogenetic position of the order Lagomorpha (rabbits, hares and allies). Nature 379, 333–335.
Response
I cannot believe that you have published this complete scientific nonsense without it being peer reviewed by the original reviewers of the paper. Surely this is the usual practice for any Letter to the Editor? If following such peer review the conclusion was that the points raised had some legitimacy then we would have gladly answered such comments. You did not afford us this opportunity and for someone who has published with JAD almost from its first volume this is extremely disappointing.
I appreciate the comment by Dr John Savory and I thank him for his time and support. However, he is not to know the nature of the individual submitting these comments. He is also not to know that this individual has been writing his scientific nonsense on behalf of the aluminium industry on every open platform on the internet. He is a troll, no more or less. I am appalled that JAD has not recognised this. He will now use your letter to support his views throughout the internet. You are allowing him to spread his misinformation through a respected source, JAD.
I also question the suggestion by the author of this internet nonsense that you, JAD, actually asked for comments on this paper. Is this true?
PubPeer or Peer Review?
In January 2020 immediately following the publication of our ‘landmark’ (as described by George Perry, Editor-in-Chief of JAD) paper on Al in brain tissue in FAD, two anonymous comments were posted on website PubPeer. This website describes itself as ‘the online journal club’. Really? An online journal club that supports anonymous comments about peer-reviewed published science. PubPeer is no more than a publishing platform for trolls. A recent review of the content of this site found that almost 90% of submitted comments were anonymous. The owners of the site stating that anonymity protects graduate students wishing to comment on published science. Really? Or perhaps it is because no one used the site before they introduced anonymity. Either way, why would any repoutable scientist use this platform for science?
The anonymous comments were made by someone calling themselves Hoya camphorifolia, a plant. An interesting choice of disguise for a troll, charged with planting seeds of doubt into any published science that does not follow the industry narrative.
We now learn that the anonymous plant Hoya camphorifolia is Abraham Al-Ahmad a ‘scientist’ working at Texas Tech University. One does wonder how this person has time to do any science bearing in mind that he has made over 2000 comments on PubPeer over the past few years. He is clearly a talented polymath as he has no reservations about commenting upon almost any subject. He has used his extensive knowledge and understanding to follow up his anonymous comments on PubPeer to submit a Letter to the Editor on our paper (and indeed all of our research on aluminium in human health).
I have no issue with any legitimate commentary on our research. In fact I welcome all informed discussion of our published research.
I do have an issue with how this Letter to the Editor has come about.
I would like George Perry to explain why in February of this year, four years after the anonymous comments were left on PubPeer, he invited an anonymous plant to write a Letter to the Editor about their issues with our paper. Is this normal practice for a journal editor? If so, why now, why not when the anonymous comments were made in January 2020.
I am then further perplexed by what exactly constitutes a Letter to the Editor? In my forty years or so in science I have always understood that Letters to the Editor about a paper published in their journal would in the first instance be peer-reviewed, commonly by the original reviewers of the published paper. If upon peer review it is decided that the Letter is worthy of publication then the authors of the paper in question are afforded an opportunity to reply to the Letter. I would like George Perry to explain why this procedure was not followed for this Letter about our paper.
I have supported JAD with my very best research almost since the first volume. I have enormous respect for George Perry, an exceptional scientist and an editor with integrity. The latter being something of a rarity these days. So, why did George Perry follow this course of action? Why did he decide to allow a known troll to publish a non-peer-reviewed unfounded comment about a paper that he himself described as a ‘landmark’ paper. George Perry has always, at least up until now, stood up for fairness and integrity in science. He has always put good science first. What or who changed this?
I have never replied to any anonymous, non-peer-reviewed comments about our research. I am not about to start doing so now. For an editor that I have always held in very high esteem to ask me to do so now is to say the very least disappointing but even more it is worrying that someone with intergrity might be so easily corrupted, probably by the power of publishers and those that control them. The aluminium industry failed in preventing science opening up their Pandora's box. We now only have to look inside to know the damage that aluminium continues to wreak on humanity.






- Comment
|